-
Courses

Courses
Choosing a course is one of the most important decisions you'll ever make! View our courses and see what our students and lecturers have to say about the courses you are interested in at the links below.
-
University Life

University Life
Each year more than 4,000 choose University of Galway as their University of choice. Find out what life at University of Galway is all about here.
-
About University of Galway

About University of Galway
Since 1845, University of Galway has been sharing the highest quality teaching and research with Ireland and the world. Find out what makes our University so special – from our distinguished history to the latest news and campus developments.
-
Colleges & Schools

Colleges & Schools
University of Galway has earned international recognition as a research-led university with a commitment to top quality teaching across a range of key areas of expertise.
-
Research & Innovation

Research & Innovation
University of Galway’s vibrant research community take on some of the most pressing challenges of our times.
-
Business & Industry

Guiding Breakthrough Research at University of Galway
We explore and facilitate commercial opportunities for the research community at University of Galway, as well as facilitating industry partnership.
-
Alumni & Friends

Alumni & Friends
There are 128,000 University of Galway alumni worldwide. Stay connected to your alumni community! Join our social networks and update your details online.
-
Community Engagement

Community Engagement
At University of Galway, we believe that the best learning takes place when you apply what you learn in a real world context. That's why many of our courses include work placements or community projects.
CHEMDiffs help
CHEMDiffs v2.20 (Sept. 2004)
Detailed chemical kinetic models can contain a very large number of species involved in exceedingly many reactions. It is very difficult to digest such datasets, so we (S¨¦bastien Rolland and John Simmie) have devised Visual Basic applications to (1) cleanup a Chemkin input mechanism file and (2) to compare two 'clean' mechanisms. Thus, these applications provide an automatic comparison of two detailed chemical kinetic mechanisms at the input level, and render what can be a laborious and error-prone procedure into, if not a pleasurable, at least a reduced stress activity.
Nota Bene: You must use a full stop "." as the decimal point and not a comma "," in your input files. And you must reset your computer via "Control Panel/Regional Options/Numbers" to Decimal Symbol="full" stop.
Installation
Unzip the downloaded CHEMDiffs.zip, and launch the file called "setup.exe". You may be required to restart your computer (recommended).
We have successfully tested the applications on a few computers in our department (mainly Windows 98, 2000 and XP). However, downloading and installing are done entirely at your own risk.
sebastien.rolland@nuigalway.ie Email us with your details if you want to be notified of updates.
After installation, your computer configuration could look like this:
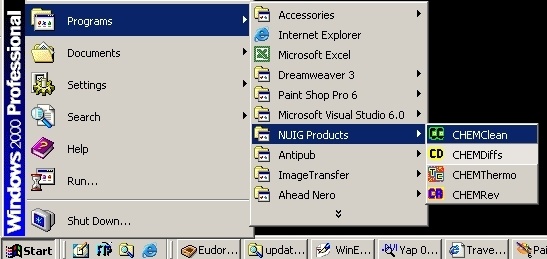
Uninstallation? Use the function "Add/Remove Programs" from your Control Panel.
How Does CHEMDiffs Work?
Two files treated successfully with CHEMClean 2.20 can be compared with CHEMDiffs 2.20.
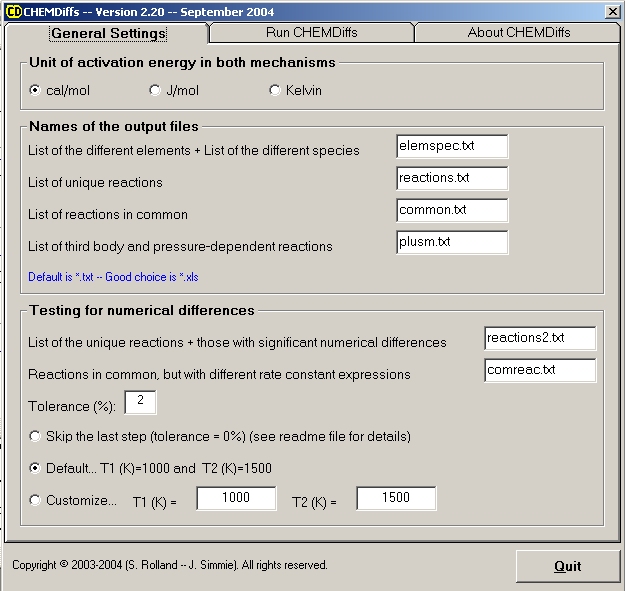
1st Step: GENERAL SETTINGS
Check if the settings of the programme are suitable to your mechanism. Default settings are as shown on the next figure.
The best option is to use the default settings, we have chosen these from our own experience of CHEMKIN files.
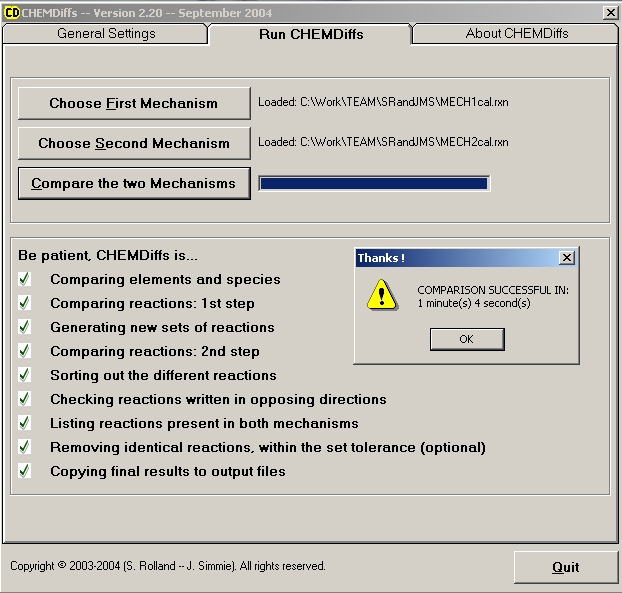
2nd Step: RUN CHEMDiffs
- Load the first clean mechanism using the button "Choose First Mechanism".
- Load the second clean mechanism using the button "Choose Second Mechanism".
CHEMDiffs then checks to see whether "END" keyword appears 3 times.
- Click on "Compare the two Mechanisms". Then relax, CHEMDiffs will do the rest. Results of comparisons will be obtained within a few minutes.
During The Automated "comparison Process"
- CHEMDiffs checks if each element present in the first file is also present in the second file and vice-versa.
- CHEMDiffs checks if each species present in the first file is also present in the second file and vice-versa.
- The reactions comparison requires many steps. Note that in the process, the programme keeps all auxiliary information data (marked with keywords LOW, HIGH, TROE, LT, REV...), relevant to a reaction. At most, 3 lines of auxiliary information below a reaction are allowed.
Warning! CHEMDiffs distinguishes third-body and pressure-dependent reactions. Thus, CH3+CH3+M < = > C2H6+M is considered different to CH3+CH3(+M) < = > C2H6(+M). These special reactions are listed in a file called by default "plusm.txt".
A rough comparison allows us to remove every reaction which appears with exactly the same format (same order of species, same rate coefficients). Two lists containing the reactions unique to each mechanism are created. Within these lists, some reactions are identical but written differently. Example:
O+HCO < = > OH+CO 3.000E+13 0.0000 0.0 (Mech #1)
must absolutely be detected as equivalent to
HCO+O < = > CO+OH 3.000E+13 0.0000 0.0 (Mech #2)
and removed from the lists.
Within these lists a large number of reactions are similar but numerically different, e.g.: H+C2H6< = >C2H5+H2 1.150E+08 1.9000 7530.0 (Mech #1) and
C2H6+H< = >C2H5+H2 1.450E+09 1.5000 7411.6 (Mech #2).
The next step evaluates how large the difference is, by calculating rate constants k at two different temperatures:
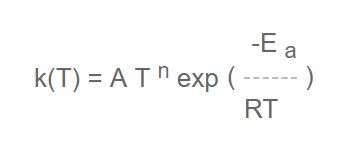
Since the numerical differences could be inconsequential, arising out of rounding off errors in the conversion from one set of units to another say, a further optional computation is performed at two user-chosen temperatures (default 1000 and 1500K) of a quantity, D, defined as:
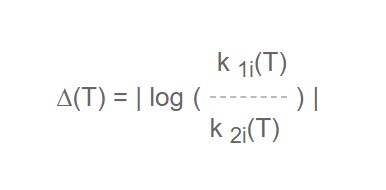
where k 1i is the rate constant in mechanism #1 for the i th reaction and k 2i is the rate constant for the same reaction in mechanism #2. Values of D of less than 2% (user-selected tolerance) at both temperatures are a signal that the rate data are essentially similar and that therefore these reactions are in common and are not truly different. They should be removed from the previous lists.
The last step sorts reactions written in the opposing direction (e.g. CH4+H < = > CH3+H2 and CH3+H2 < = > CH4+H). You will need to use another software called CHEMRev, to know if they are numerically equivalent.
When the programme ends a message box pops up, and states how long it took to do the comparison.
Exceptionally, an error message box may appear since the programme also carries out a number of logical tests. These include the computation of:
CE = |(NE1 - NE1P) - (NE2 - NE_2P)|
CS = |(NS1 - NS1P) - (NS_2 - NS_2P)|
CR = |(NR1 - NR1P) - (NR_2 - NR_2P)| where
NE 1, NS 1, NR 1 are respectively the number of elements, species, reactions in MECH #1
NE 2, NS 2, NR 2 are respectively the number of elements, species, reactions in MECH #2
NE 1P, NS 1P, NR 1P are respectively the number of elements, species, reactions in MECH #1 and not in MECH #2
NE 2P, NS 2P, NR 2P are respectively the number of elements, species, reactions in MECH #2 and not in MECH #1
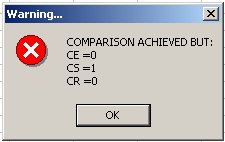
This checks if the number of elements/species/reactions in common between the two mechanisms is the same. We should have CE = CS = CR = 0. The example shown above warns us that there is a problem in the species information (CE = 0, CS="1," CR="0)." This can happen if a species is declared twice in a mechanism and only once in another one.
Output Files
CHEMDiffs supplies 7 final output files. Default names files are:
 "elemspec.txt" lists the missing elements and species.
"elemspec.txt" lists the missing elements and species.
 "reactions.txt" draws up the list of reactions which are present in MECH #1 but not in the second one + the list of reactions which are present in MECH #2 but not in the first one.
"reactions.txt" draws up the list of reactions which are present in MECH #1 but not in the second one + the list of reactions which are present in MECH #2 but not in the first one.
 "reactions2.txt" draws up the list of reactions which are present in MECH #1 but not in the second one + the list of reactions which are present in MECH #2 but not in the first one. Also are included in this file, the reactions that are common to both mechanisms but with significantly different rate constants expressions.
"reactions2.txt" draws up the list of reactions which are present in MECH #1 but not in the second one + the list of reactions which are present in MECH #2 but not in the first one. Also are included in this file, the reactions that are common to both mechanisms but with significantly different rate constants expressions.
 "common.txt" presents the list of reactions that appear as written in both mechanisms and have identical rate constants expressions + the list of reactions that appear in both mechanisms but with different rate constants expressions (even slightly) + the list of reactions that are written in opposing directions in the opposing mechanisms + the list of reactions that are identical within the set tolerance of 2%. Note that the symbol "(#)" appears at the beginning of each equation, whose reactants and products order, are different. The symbol "(###)" is a tag, that will be used by another of our software CHEMRev, to treat the reactions written in opposing directions in the opposing mechanisms.
"common.txt" presents the list of reactions that appear as written in both mechanisms and have identical rate constants expressions + the list of reactions that appear in both mechanisms but with different rate constants expressions (even slightly) + the list of reactions that are written in opposing directions in the opposing mechanisms + the list of reactions that are identical within the set tolerance of 2%. Note that the symbol "(#)" appears at the beginning of each equation, whose reactants and products order, are different. The symbol "(###)" is a tag, that will be used by another of our software CHEMRev, to treat the reactions written in opposing directions in the opposing mechanisms.
 "plusm.txt" lists the third-body and pressure-dependent reactions.
"plusm.txt" lists the third-body and pressure-dependent reactions.
 "comreac.txt" presents the list of reactions similar to the two mechanisms but with significantly different rate constants expressions (D(T) <2%). Note that the symbol "(#)" appears at the beginning of each equation, whose numbers, reactants and products order, are different.
"comreac.txt" presents the list of reactions similar to the two mechanisms but with significantly different rate constants expressions (D(T) <2%). Note that the symbol "(#)" appears at the beginning of each equation, whose numbers, reactants and products order, are different.
 Finally, an output file called "Summary.txt", gives the abstract of the comparison. This file specifies the number of species and reactions (specifying the number of times the word "DUPLICATE" appears), and the number of species and reactions which are not present in each of 2 kinetic mechanisms.
Finally, an output file called "Summary.txt", gives the abstract of the comparison. This file specifies the number of species and reactions (specifying the number of times the word "DUPLICATE" appears), and the number of species and reactions which are not present in each of 2 kinetic mechanisms.
 In order to help read the CHEMDiffs output files, we advise to import them from any existing tabler. The extension *.txt has been chosen by default but *.xls is perfectly accepted for common.txt (common.xls), comreac.txt (comreac.xls), reactions.txt (reactions.xls), reactions2.txt (reactions2.xls).
In order to help read the CHEMDiffs output files, we advise to import them from any existing tabler. The extension *.txt has been chosen by default but *.xls is perfectly accepted for common.txt (common.xls), comreac.txt (comreac.xls), reactions.txt (reactions.xls), reactions2.txt (reactions2.xls).
Limitations
CHEMDiffs uses tablers provided by Visual BasiccircledR, limited by a certain number of rows. Unfortunately, we are not able to predict from what number of reactions, the programme may crash. We recommend that the number of lines in you mechanism does not exceed 14580 lines. Contact us if you encounter this issue.
CHEMDiffs doesn't compare non-elementary reactions, that are placed at the end of the cleaned mechanism. Reactions such as A+B+C = 2D+F+2E are supported but note that CHEMDiffs will treat "2D" and "2E" as species. This particular reaction will be considered different from A+B+C = D+D+F+2E.
"DUPLICATE" reactions in the files "common.txt" and "comreac.txt":
|
HO2+HO2 = O2+H2O2 1.300E11 0.0000 -1630.0 (from [Mech # 1])
|
(1)
|
|
DUPLICATE
|
|
|
HO2+HO2 = O2+H2O2 4.200E14 0.0000 12000.0 (from [Mech # 1])
|
(2)
|
|
DUPLICATE
|
|
|
HO2+HO2 = H2O2+O2 1.320E11 0.0000 -1630.0 (from [Mech # 2])
|
(3)
|
|
DUPLICATE
|
|
|
HO2+HO2 = H2O2+O2 4.220E12 0.0000 11983.7 (from [Mech # 2])
|
(4)
|
|
DUPLICATE
|
|















